
Generate Random Samples from Multivariate Normal Distribution
Source:R/utility_rmvnorm.R
rmvnorm.RdIn \(\mathbf{R}^p\), random samples are drawn $$X_1,X_2,\ldots,X_n~ \sim ~ \mathcal{N}(\mu, \Sigma)$$ where \(\mu \in \mathbf{R}^p\) is a mean vector and \(\Sigma \in \textrm{SPD}(p)\) is a positive definite covariance matrix.
Value
either (1) a length-\(p\) vector (\(n=1\)) or (2) an \((n\times p)\) matrix where rows are random samples.
Examples
#-------------------------------------------------------------------
# Generate Random Data and Compare with Empirical Covariances
#
# In R^5 with zero mean and diagonal covariance,
# generate 100 and 200 observations and compute MLE covariance.
#-------------------------------------------------------------------
## GENERATE DATA
mymu = rep(0,5)
mysig = diag(5)
## MLE FOR COVARIANCE
smat1 = stats::cov(rmvnorm(n=100, mymu, mysig))
smat2 = stats::cov(rmvnorm(n=200, mymu, mysig))
## VISUALIZE
opar <- par(no.readonly=TRUE)
par(mfrow=c(1,3), pty="s")
image(mysig[,5:1], axes=FALSE, main="true covariance")
image(smat1[,5:1], axes=FALSE, main="empirical cov with n=100")
image(smat2[,5:1], axes=FALSE, main="empirical cov with n=200")
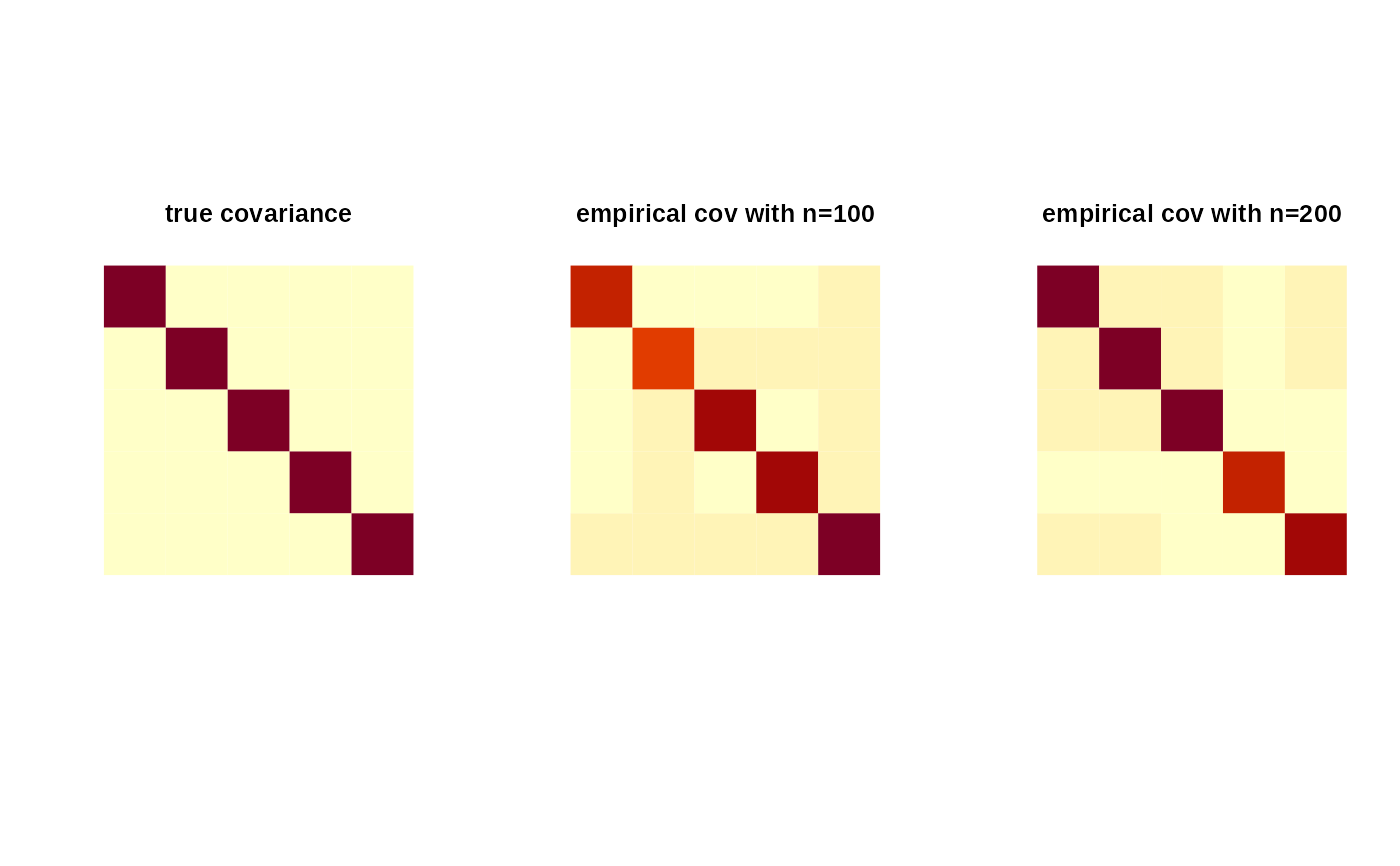 par(opar)
par(opar)