Given \(N\) observations \(X_1, X_2, \ldots, X_N \in \mathcal{M}\), perform k-means++ clustering algorithm using pairwise distances. The algorithm was originally designed as an efficient initialization method for k-means algorithm.
Usage
riem.kmeanspp(riemobj, k = 2, geometry = c("intrinsic", "extrinsic"))Value
a named list containing
- centers
a length-\(k\) vector of sampled centers' indices.
- cluster
a length-\(N\) vector of class labels (from \(1:k\)).
References
Arthur D, Vassilvitskii S (2007). “K-Means++: The advantages of careful seeding.” In Proceedings of the eighteenth annual ACM-SIAM symposium on discrete algorithms, SODA '07, 1027–1035. ISBN 978-0-89871-624-5, Number of pages: 9 Place: New Orleans, Louisiana.
Examples
#-------------------------------------------------------------------
# Example on Sphere : a dataset with three types
#
# class 1 : 10 perturbed data points near (1,0,0) on S^2 in R^3
# class 2 : 10 perturbed data points near (0,1,0) on S^2 in R^3
# class 3 : 10 perturbed data points near (0,0,1) on S^2 in R^3
#-------------------------------------------------------------------
## GENERATE DATA
mydata = list()
for (i in 1:10){
tgt = c(1, stats::rnorm(2, sd=0.1))
mydata[[i]] = tgt/sqrt(sum(tgt^2))
}
for (i in 11:20){
tgt = c(rnorm(1,sd=0.1),1,rnorm(1,sd=0.1))
mydata[[i]] = tgt/sqrt(sum(tgt^2))
}
for (i in 21:30){
tgt = c(stats::rnorm(2, sd=0.1), 1)
mydata[[i]] = tgt/sqrt(sum(tgt^2))
}
myriem = wrap.sphere(mydata)
mylabs = rep(c(1,2,3), each=10)
## K-MEANS++ WITH K=2,3,4
clust2 = riem.kmeanspp(myriem, k=2)
clust3 = riem.kmeanspp(myriem, k=3)
clust4 = riem.kmeanspp(myriem, k=4)
## MDS FOR VISUALIZATION
mds2d = riem.mds(myriem, ndim=2)$embed
## VISUALIZE
opar <- par(no.readonly=TRUE)
par(mfrow=c(2,2), pty="s")
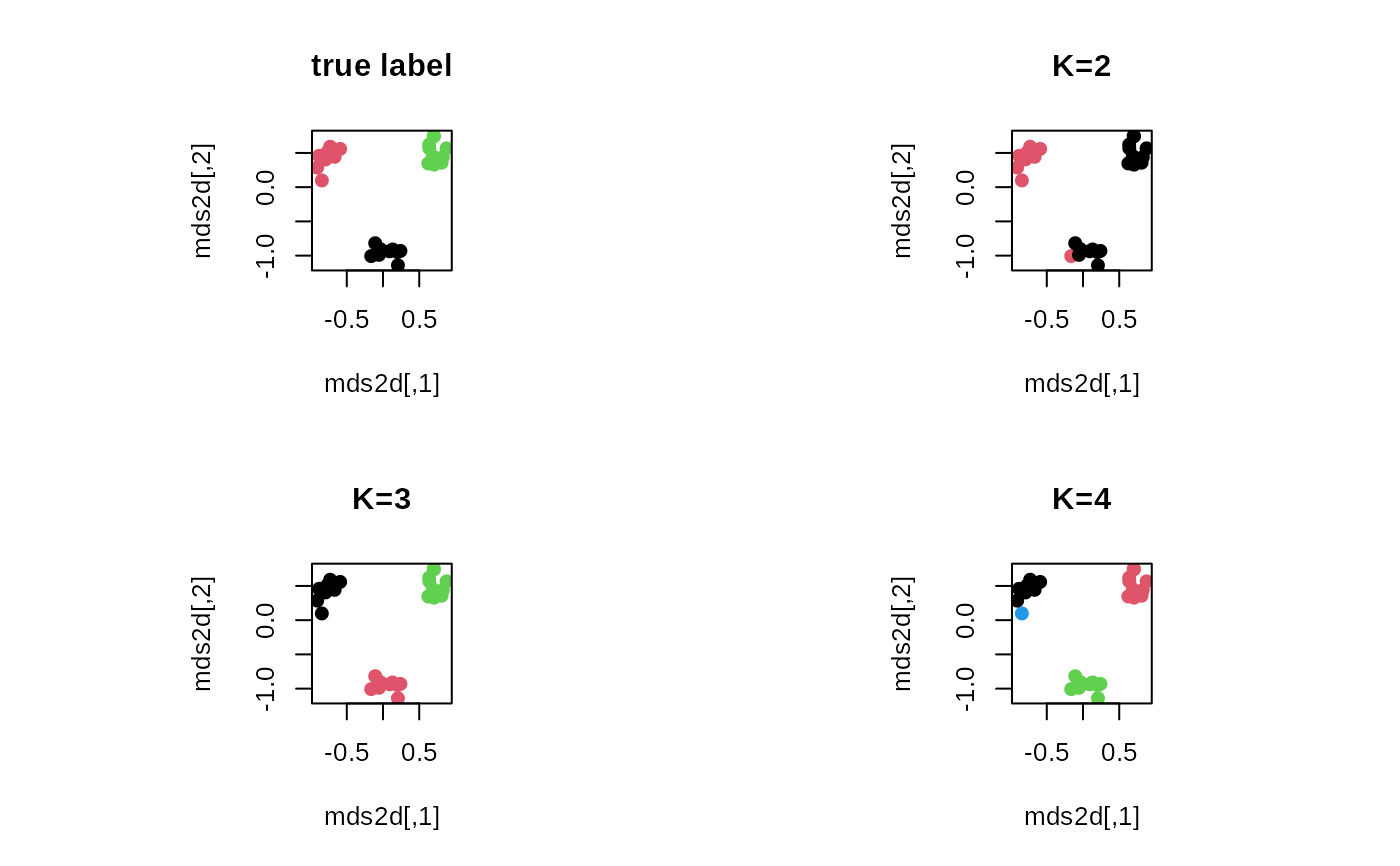
plot(mds2d, pch=19, main="true label", col=mylabs)
plot(mds2d, pch=19, main="K=2", col=clust2$cluster)
plot(mds2d, pch=19, main="K=3", col=clust3$cluster)
plot(mds2d, pch=19, main="K=4", col=clust4$cluster)
 par(opar)
par(opar)
