For a hypersphere \(\mathcal{S}^{p-1}\) in \(\mathbf{R}^p\), Angular Central Gaussian (ACG) distribution \(ACG_p (A)\) is defined via a density $$f(x\vert A) = |A|^{-1/2} (x^\top A^{-1} x)^{-p/2}$$ with respect to the uniform measure on \(\mathcal{S}^{p-1}\) and \(A\) is a symmetric positive-definite matrix. Since \(f(x\vert A) = f(-x\vert A)\), it can also be used as an axial distribution on real projective space, which is unit sphere modulo \(\lbrace{+1,-1\rbrace}\). One constraint we follow is that \(f(x\vert A) = f(x\vert cA)\) for \(c > 0\) in that we use a normalized version for numerical stability by restricting \(tr(A)=p\).
Arguments
- datalist
a list of length-\(p\) unit-norm vectors.
- A
a \((p\times p)\) symmetric positive-definite matrix.
- n
the number of samples to be generated.
- ...
extra parameters for computations, including
- maxiter
maximum number of iterations to be run (default:50).
- eps
tolerance level for stopping criterion (default: 1e-5).
Value
dacg gives a vector of evaluated densities given samples. racg generates
unit-norm vectors in \(\mathbf{R}^p\) wrapped in a list. mle.acg estimates
the SPD matrix \(A\).
References
Tyler DE (1987). “Statistical analysis for the angular central Gaussian distribution on the sphere.” Biometrika, 74(3), 579–589. ISSN 0006-3444, 1464-3510.
Mardia KV, Jupp PE (eds.) (1999). Directional Statistics, Wiley Series in Probability and Statistics. John Wiley \& Sons, Inc., Hoboken, NJ, USA. ISBN 978-0-470-31697-9 978-0-471-95333-3.
Examples
# -------------------------------------------------------------------
# Example with Angular Central Gaussian Distribution
#
# Given a fixed A, generate samples and estimate A via ML.
# -------------------------------------------------------------------
## GENERATE AND MLE in R^5
# Generate data
Atrue = diag(5) # true SPD matrix
sam1 = racg(50, Atrue) # random samples
sam2 = racg(100, Atrue)
# MLE
Amle1 = mle.acg(sam1)
Amle2 = mle.acg(sam2)
# Visualize
opar <- par(no.readonly=TRUE)
par(mfrow=c(1,3), pty="s")
image(Atrue[,5:1], axes=FALSE, main="true SPD")
image(Amle1[,5:1], axes=FALSE, main="MLE with n=50")
image(Amle2[,5:1], axes=FALSE, main="MLE with n=100")
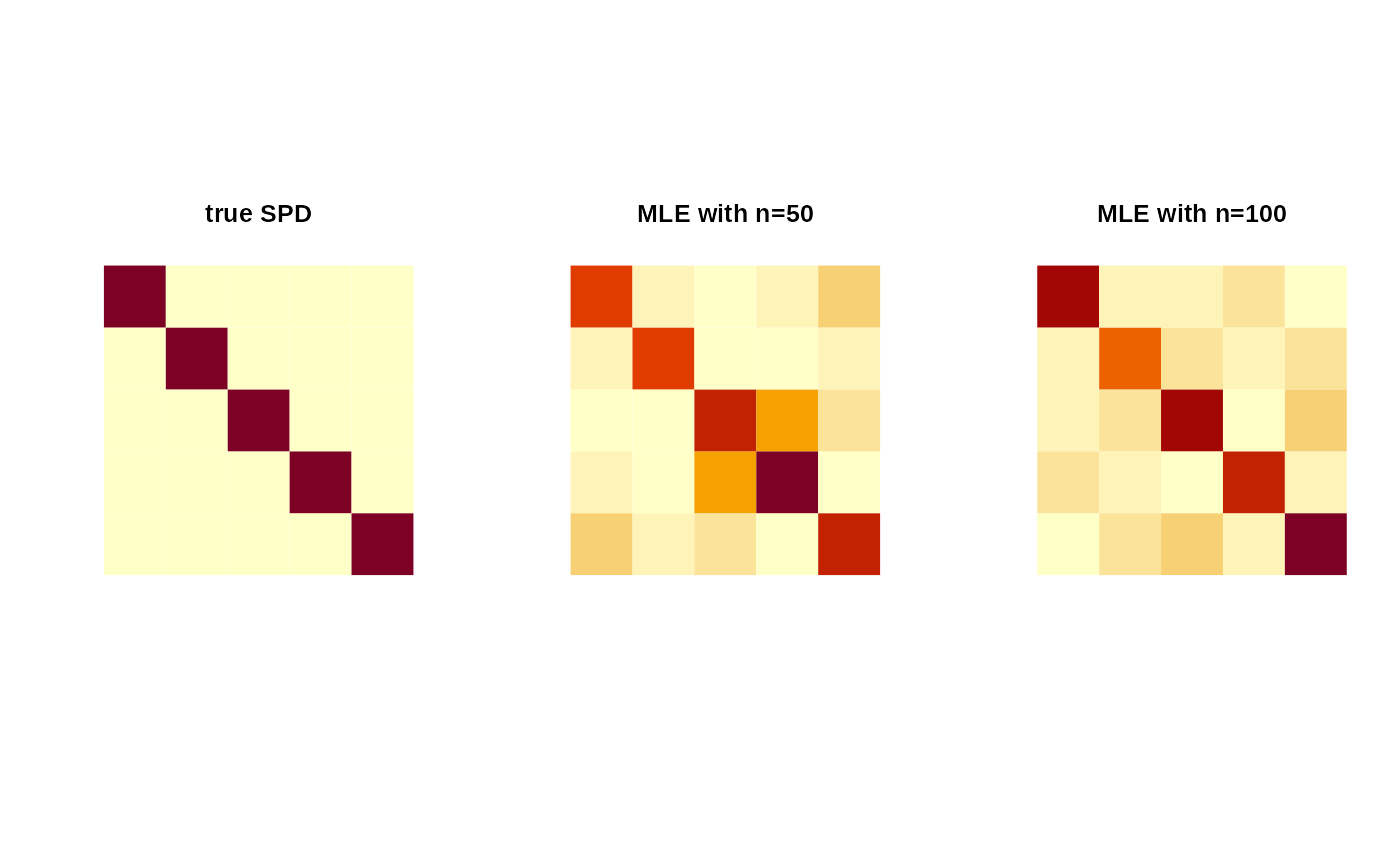 par(opar)
par(opar)
