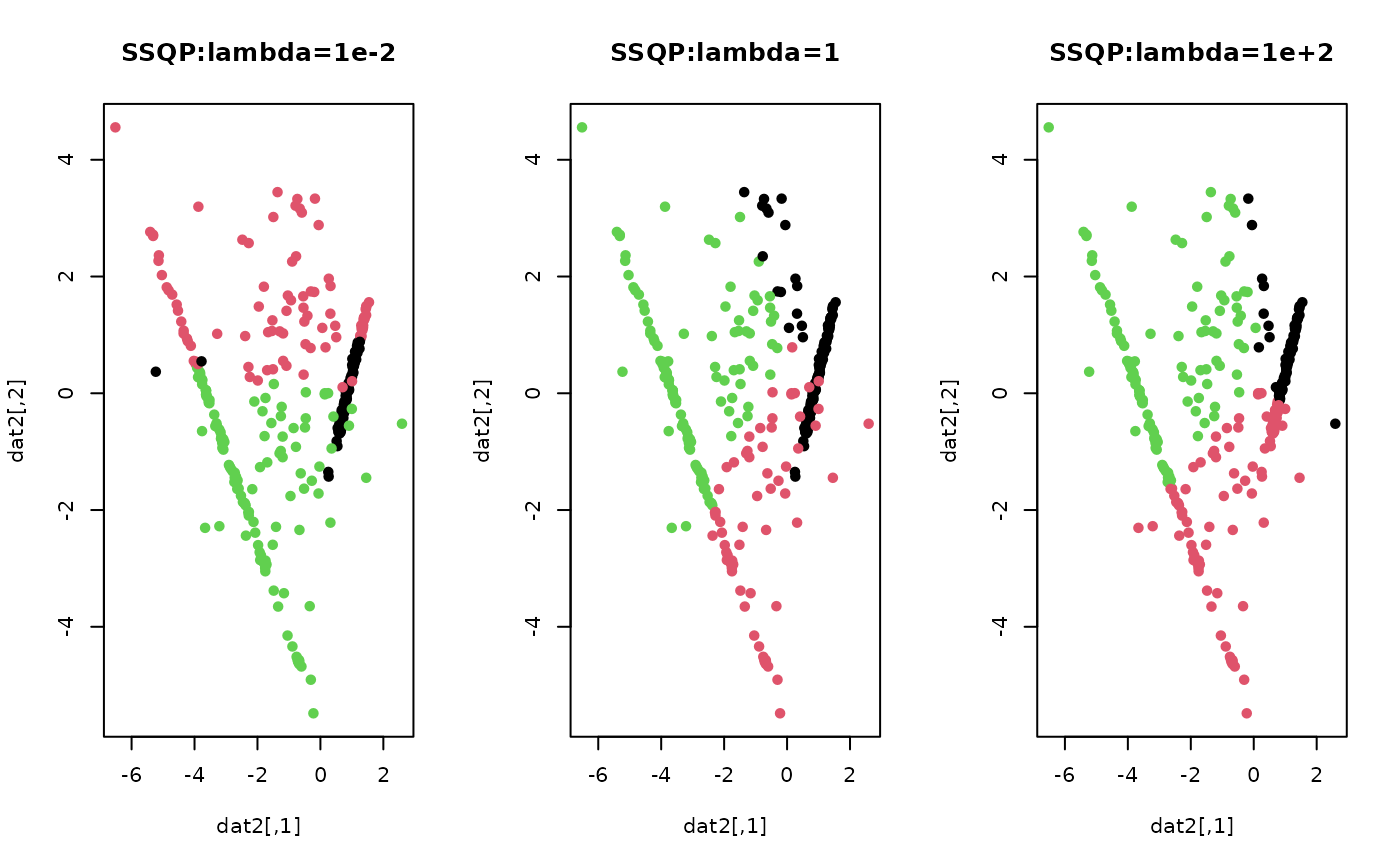Subspace Segmentation via Quadratic Programming (SSQP) solves the following problem $$\textrm{min}_Z \|X-XZ\|_F^2 + \lambda \|Z^\top Z\|_1 \textrm{ such that }diag(Z)=0,~Z\leq 0$$ where \(X\in\mathbf{R}^{p\times n}\) is a column-stacked data matrix. The computed \(Z^*\) is used as an affinity matrix for spectral clustering.
SSQP(data, k = 2, lambda = 1e-05, ...)
Arguments
| data | an \((n\times p)\) matrix of row-stacked observations. |
|---|---|
| k | the number of clusters (default: 2). |
| lambda | regularization parameter (default: 1e-5). |
| ... | extra parameters for the gradient descent algorithm including
|
Value
a named list of S3 class T4cluster containing
- cluster
a length-\(n\) vector of class labels (from \(1:k\)).
- algorithm
name of the algorithm.
References
Wang S, Yuan X, Yao T, Yan S, Shen J (2011). “Efficient Subspace Segmentation via Quadratic Programming.” In Proceedings of the Twenty-Fifth AAAI Conference on Artificial Intelligence, AAAI'11, 519--524.
Examples
# \donttest{ ## generate a toy example set.seed(10) tester = genLP(n=100, nl=2, np=1, iso.var=0.1) data = tester$data label = tester$class ## do PCA for data reduction proj = base::eigen(stats::cov(data))$vectors[,1:2] dat2 = data%*%proj ## run SSQP for k=3 with different lambda values out1 = SSQP(data, k=3, lambda=1e-2) out2 = SSQP(data, k=3, lambda=1) out3 = SSQP(data, k=3, lambda=1e+2) ## extract label information lab1 = out1$cluster lab2 = out2$cluster lab3 = out3$cluster ## visualize opar <- par(no.readonly=TRUE) par(mfrow=c(1,3)) plot(dat2, pch=19, cex=0.9, col=lab1, main="SSQP:lambda=1e-2") plot(dat2, pch=19, cex=0.9, col=lab2, main="SSQP:lambda=1") plot(dat2, pch=19, cex=0.9, col=lab3, main="SSQP:lambda=1e+2")