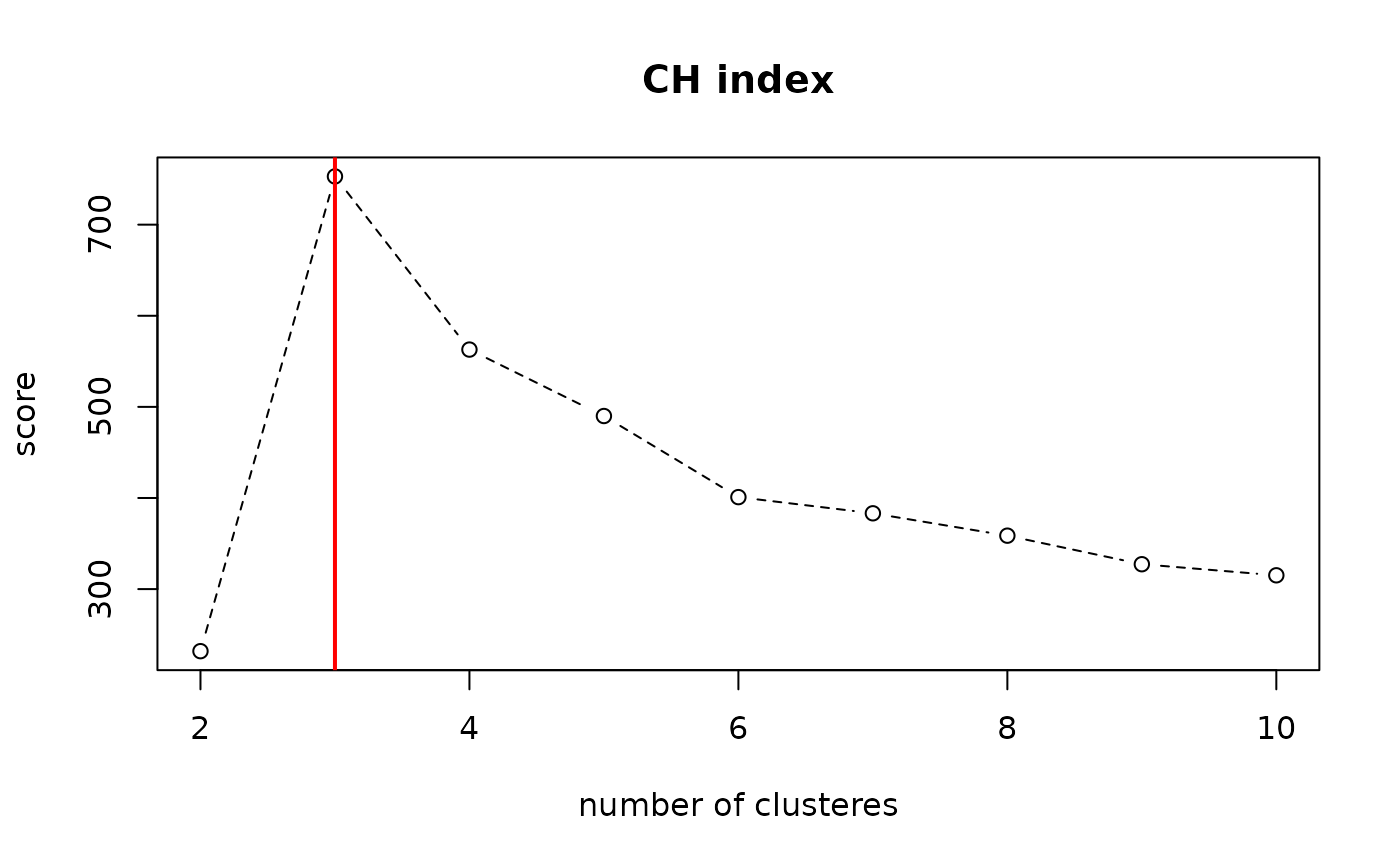
Calinski and Harabasz index. Higher score means a good clustering.
quality.CH(data, cluster)
Arguments
| data | an \((n\times p)\) matrix of row-stacked observations. |
|---|---|
| cluster | a length-\(n\) vector of class labels (from \(1:k\)). |
Value
an index value.
Examples
# \donttest{ # ------------------------------------------------------------- # clustering validity check with 3 Gaussians # ------------------------------------------------------------- ## PREPARE X1 = matrix(rnorm(30*3, mean=-5), ncol=3) X2 = matrix(rnorm(30*3), ncol=3) X3 = matrix(rnorm(30*3, mean=5), ncol=3) XX = rbind(X1, X2, X3) ## CLUSTERING WITH DIFFERENT K VALUES & COMPUTE QUALITY INDICES vec_k = 2:10 vec_cl = rep(0, 9) for (i in 1:length(vec_k)){ cl_now = T4cluster::kmeans(XX, k=vec_k[i])$cluster vec_cl[i] = quality.CH(XX, cl_now) } ## VISUALIZE opar <- par(no.readonly=TRUE) plot(vec_k, vec_cl, type="b", lty=2, xlab="number of clusteres", ylab="score", main="CH index")