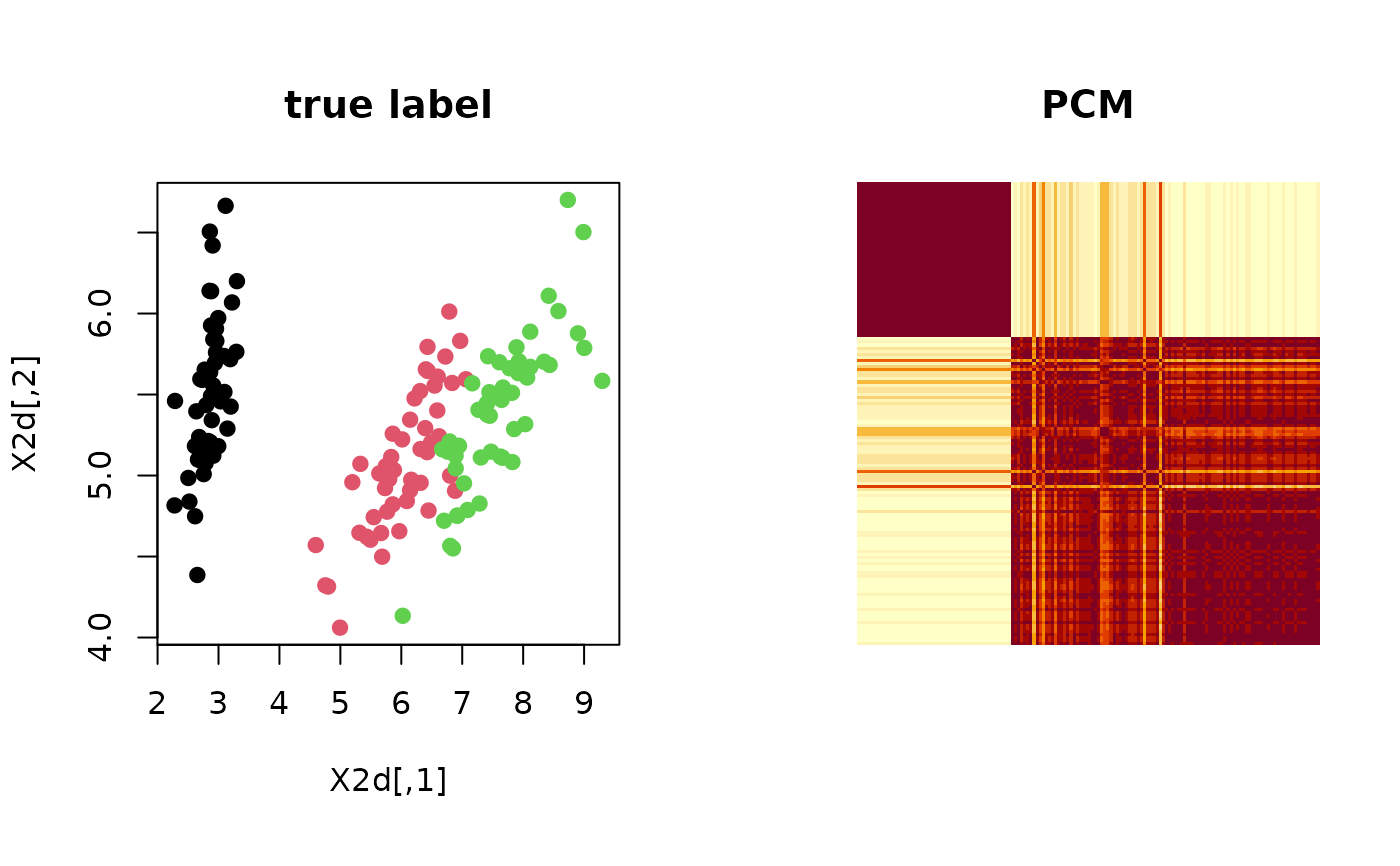Let clustering be a label from data of \(N\) observations and suppose we are given \(M\) such labels. Co-occurrent matrix counts the number of events where two observations \(X_i\) and \(X_j\) belong to the same category/class. PCM serves as a measure of uncertainty embedded in any algorithms with non-deterministic components.
pcm(partitions)
Arguments
| partitions | partitions can be provided in either (1) an \((M\times N)\) matrix where each row is a clustering for \(N\) objects, or (2) a length-\(M\) list of length-\(N\) clustering labels. |
|---|
Value
an \((N\times N)\) matrix, whose elements \((i,j)\) are counts for how many times observations \(i\) and \(j\) belong to the same cluster, ranging from \(0\) to \(M\).
See also
Examples
# ------------------------------------------------------------- # PSM with 'iris' dataset + k-means++ # ------------------------------------------------------------- ## PREPARE WITH SUBSET OF DATA data(iris) X = as.matrix(iris[,1:4]) lab = as.integer(as.factor(iris[,5])) ## EMBEDDING WITH PCA X2d = Rdimtools::do.pca(X, ndim=2)$Y ## RUN K-MEANS++ 100 TIMES partitions = list() for (i in 1:100){ partitions[[i]] = kmeanspp(X)$cluster } ## COMPUTE PCM iris.pcm = pcm(partitions) ## VISUALIZATION opar <- par(no.readonly=TRUE) par(mfrow=c(1,2), pty="s") plot(X2d, col=lab, pch=19, main="true label") image(iris.pcm[,150:1], axes=FALSE, main="PCM")